Researchers from Universidad Autónoma de Madrid published a new editorial in Oncotarget detailing a proof-of-principle experiment to prevent B-cell acute lymphoblastic leukemia (B-ALL).

The Trending With Impact series highlights Oncotarget publications attracting higher visibility among readers around the world online, in the news, and on social media—beyond normal readership levels. Look for future science news about the latest trending publications here, and at Oncotarget.com.
—
Childhood leukemia is a devastating disease that affects thousands of children every year. Despite significant advancements in the field of pediatric oncology, childhood leukemia remains a major cause of morbidity and mortality in children, with B-cell acute lymphoblastic leukemia (B-ALL) being the most common form.
Some cases of childhood B-ALL arise from congenital mutations that lead to a silent population of pre-leukemic cells. These cells at some point are triggered by a catalyst (possibly by delayed exposure to a common infection), acquire additional genetic mutations and ultimately develop into B-ALL. However, researchers have yet to fully understand how to target these pre-leukemic cells to prevent B-ALL.
In a new editorial paper, researchers César Cobaleda, Manuel Ramírez-Orellana, Carolina Vicente-Dueñas, Andreas Weiss, Kim E. Nichols, and Isidro Sánchez-García from Universidad Autónoma de Madrid discuss a novel method of targeting pre-leukemic cells in practice. On March 11, 2023, the team published their editorial in Oncotarget, entitled, “Proof-of-principle: targeted childhood leukemia prevention.”
“[…] one would have to find a way to specifically target these preleukemic cells. Recently, a mouse model recapitulating the phenotype of a leukemia-predisposition syndrome has allowed us to carry out a proof-of-principle experiment to achieve this very goal.”
The Study
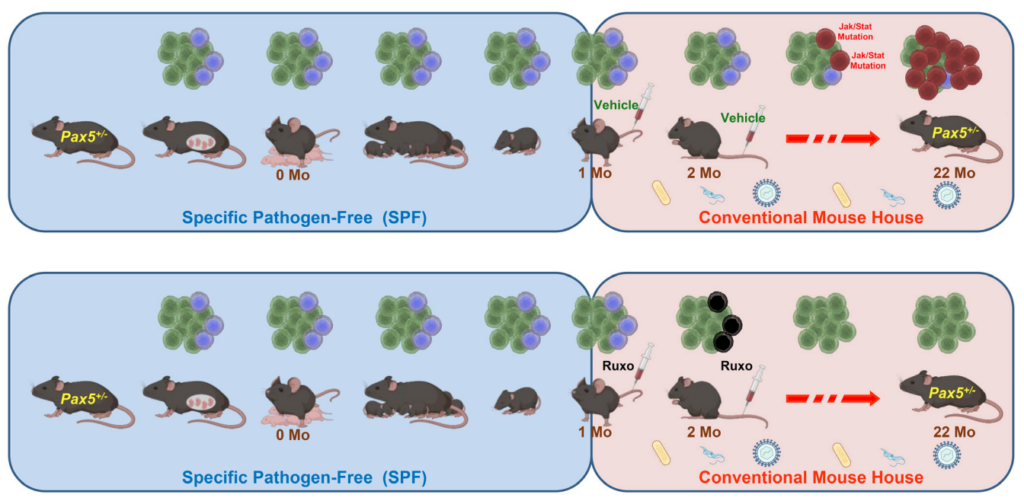
Pax5 is a protein that plays a critical role in the development of white blood cells that produce antibodies to fight infections, B cells. A Pax5 mutation or deficiency can lead to a disruption in the development of B cells and compromise the immune system’s ability to fight off infections. Pax5+/- refers to an individual or organism that has one functional copy of the Pax5 gene and one non-functional copy. This condition is also known as heterozygosity.
“Children carrying heterozygous mutations affecting the B-cell master regulator gene PAX5 are predisposed to develop B-ALL; similarly, 25% of heterozygous Pax5+/− mice develop leukemia, but only after experiencing an immune stress, such as exposure to infection [2–4].”
In their recent study, the researchers used Pax5+/− mice (a mouse model carrying a leukemia-predisposition syndrome) to evaluate whether in vivo treatment with ruxolitinib, a Jak1/2 inhibitor, administered early in life is capable of killing pre-leukemic cells and preventing the development of acute leukemia. They found that treatment with ruxolitinib led to the disappearance of B-cell progenitors in Pax5+/-, but not in wild-type (WT), mice. When both experimental Pax5+/- and control WT animals were fed with ruxolitinib-containing chow for 14 or 28 days and then exposed to common mouse pathogens, the animals treated with ruxolitinib for 28 days showed a significant 90% reduction in the incidence of B-ALL compared to untreated mice or animals treated only for 14 days.
Ultra-deep sequencing studies of Pax5+/- mice showed that ruxolitinib acts by eliminating predisposed preleukemic B cells before the second “hit” (or catalyst) that leads to their descent into B-ALL. These findings suggest that an analogous approach could be used to prevent the development of B-ALL in children who carry germline mutations that predispose them to this disease.
“It is becoming increasingly clear that the existence of latent pretumoral cells is common to many types of both hematologic and solid cancers [7]; therefore, the concept described here could be considered a proof-of-principle strategy for the development of similar prophylactic approaches to prevent the progression of other malignancies.”
Summary & Conclusion
Childhood leukemia, specifically B-cell acute lymphoblastic leukemia, remains a significant cause of morbidity and mortality in children, despite advancements in pediatric oncology. The pre-leukemic cells that predispose children to B-ALL are not fully understood, making it challenging to prevent the disease. However, the recent study by Cobaleda C, et al. offers new hope. The proof-of-principle experiment demonstrates that early treatment with ruxolitinib can eliminate pre-leukemic cells and significantly reduce the incidence of B-ALL in the Pax5+/- mouse model.
Their findings suggest that a similar approach could be used to prevent B-ALL in children with germline mutations that predispose them to this disease. The concept described in this study could also serve as a strategy for developing prophylactic approaches to prevent the progression of other malignancies that share the existence of latent pretumoral cells. While further research is necessary, this study offers new possibilities in preventing childhood leukemia and improving the outcomes for children at risk.
“Still, some aspects remain unclear. For example, why do the majority of genetically predisposed animals (and most genetically predisposed children) not develop leukemia and stay healthy? In addition, what are the mechanisms by which environmental factors such as infection promote the acquisition of secondary mutations leading to malignant progression of preleukemic cells? These and other important questions still need to be answered if we are to fully understand and avert the appearance of B-ALL.”
Click here to read the full editorial published in Oncotarget.
ONCOTARGET VIDEOS: YouTube | LabTube | Oncotarget.com
—
Oncotarget is an open-access, peer-reviewed journal that has published primarily oncology-focused research papers since 2010. These papers are available to readers (at no cost and free of subscription barriers) in a continuous publishing format at Oncotarget.com. Oncotarget is indexed/archived on MEDLINE / PMC / PubMed.
Click here to subscribe to Oncotarget publication updates.
For media inquiries, please contact media@impactjournals.com.