Researchers from Astellas Pharma Inc. investigated the efficacy of a novel antibody drug conjugate combined with venetoclax and azacitidine in a mouse model of acute myeloid leukemia.

The average age of patients with acute myeloid leukemia (AML) is 67 years old. Older adults generally have a lower tolerance for treatments that exhibit high off-target toxicity. Additionally, chemotherapy-relapsed or -refractory (R/R) AML patients are often at an advanced stage of disease and are therefore more likely to have comorbidities that may reduce their tolerance for harsh treatments.
Thus, pharmaceutical AML drugs with high efficacy and low toxicity are in high demand. Antibody drug conjugates (ADCs) are emerging as promising therapeutic approaches to more safely treat hematological malignancies by reducing side effects. ADCs are designed to decrease damage to healthy tissues by specifically targeting tumor-associated antigens attached to cancer cells.
“Antibody drug conjugates (ADC) are one of the modalities that aims to dissociate drug efficacy from toxicity. ADC consists of three components: antibody specific for tumor associated antigen, drug linker and cytotoxic payload.”
Astellas Pharma
Recently, researchers from Astellas Pharma Inc. (a pharmaceutical company in Japan) developed ASP1235—a novel ADC that targets Fms-like tyrosine kinase 3 (FLT3). In more than 90% of AML patients, FLT3 is overexpressed on leukemic blasts. ASP1235 is designed to target FLT3-positive leukemia cells and deliver the cytotoxic drug payload to these cells. However, this drug alone was found to have only a mild effect on AML cells, prompting researchers to assess the efficacy of ASP1235 in combination with other drugs.
In a new study, Astellas Pharma researchers Hirofumi Tsuzuki, Tatsuya Kawase, Taisuke Nakazawa, Masamichi Mori, and Taku Yoshida investigated the efficacy of ASP1235 combined with venetoclax (an anti-apoptotic agent) and azacitidine (a DNA methyltransferase inhibitor) in an experimental mouse model of AML. Their research paper was published in Oncotarget on December 20, 2022, and entitled, “Anti-tumor effect of antibody drug conjugate ASP1235 targeting Fms-like tyrosine kinase 3 with venetoclax plus azacitidine in an acute myeloid leukemia xenograft mouse model.”
“In this study, we sought to evaluate the therapeutic effect of ASP1235 in combination with venetoclax plus azacitidine, a novel standard-of-care treatment for elderly AML patients, in ASP1235 poor sensitive AML cells.”
The Study
The researchers first aimed to determine an AML cell line that was only partially sensitive to ASP1235 monotherapy. They determined the THP-1 cell line was appropriate for further investigation. They compared FLT3 and Bcl-2 expression levels in THP-1 cells with primary leukemic cells from chemotherapy R/R AML patients to consider the clinical relevance of each. In THP-1 cells, the expression levels of FLT3 and Bcl-2 were found to be clinically relevant.
“It has been reported that the proportion of patients showing high Bcl-2 expression was greater in chemotherapy R/R AML patients compared to that in newly diagnosed patients [4]. Thus, we investigated the expression levels of Bcl-2 together with FLT3 to further consider the relevance of THP-1 cells for evaluation on the combination treatment with venetoclax.”
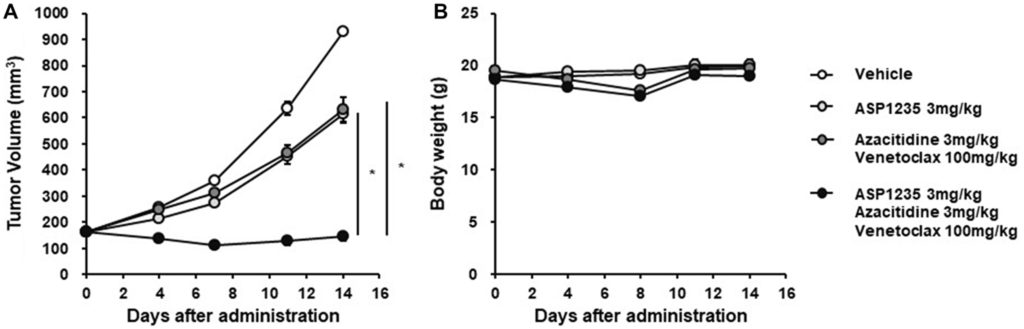
To confirm their in vitro findings, they used a THP-1 xenograft mouse model for in vivo investigation of ASP1235 sensitivity. Their findings indicated that the THP-1 cell was a partially sensitive preclinical model to ASP1235. Next, the researchers evaluated the in vivo efficacy of ASP1235 in combination with venetoclax plus azacitidine using the THP-1 xenograft mouse model. The results showed that the combination therapy induced a significant reduction in tumor size compared to ASP1235 monotherapy and the other two drugs alone. This suggests that ASP1235 has an enhanced anti-tumor effect in combination with venetoclax and azacitidine.
“Consistent with in vitro observations in Figure 4, triple combination treatment with ASP1235, venetoclax and azacitidine induced tumor regression, and the anti-tumor effect of the triple combination was much stronger than that of ASP1235 single agent or venetoclax plus azacitidine without obvious body weight loss (Figure 5).”
Conclusions
The findings of this study suggest that the combination therapy of ASP1235, venetoclax and azacitidine can be an effective treatment option for elderly patients or patients with chemotherapy R/R AML. This combination therapy induced a significant reduction in xenograft tumors in the THP-1 mouse model, suggesting that it may be a promising therapeutic approach for AML patients. Further clinical trials are needed to confirm these results.
“In conclusion, the triple combination treatment of ASP1235, venetoclax and azacitidine has the potential to benefit AML patients, and there is a possibility to expect the combination effect of ASP1235 and venetoclax regimen in FLT3 positive cancers beyond AML.”
Click here to read the full research paper published by Oncotarget.
ONCOTARGET VIDEOS: YouTube | LabTube | Oncotarget.com
—
Oncotarget is an open-access journal that publishes primarily oncology-focused research papers in a continuous publishing format. These papers are available at no cost to readers on Oncotarget.com. Open-access journals have the power to benefit humanity from the inside out by rapidly disseminating information that may be freely shared with researchers, colleagues, family, and friends around the world.
For media inquiries, please contact media@impactjournals.com.